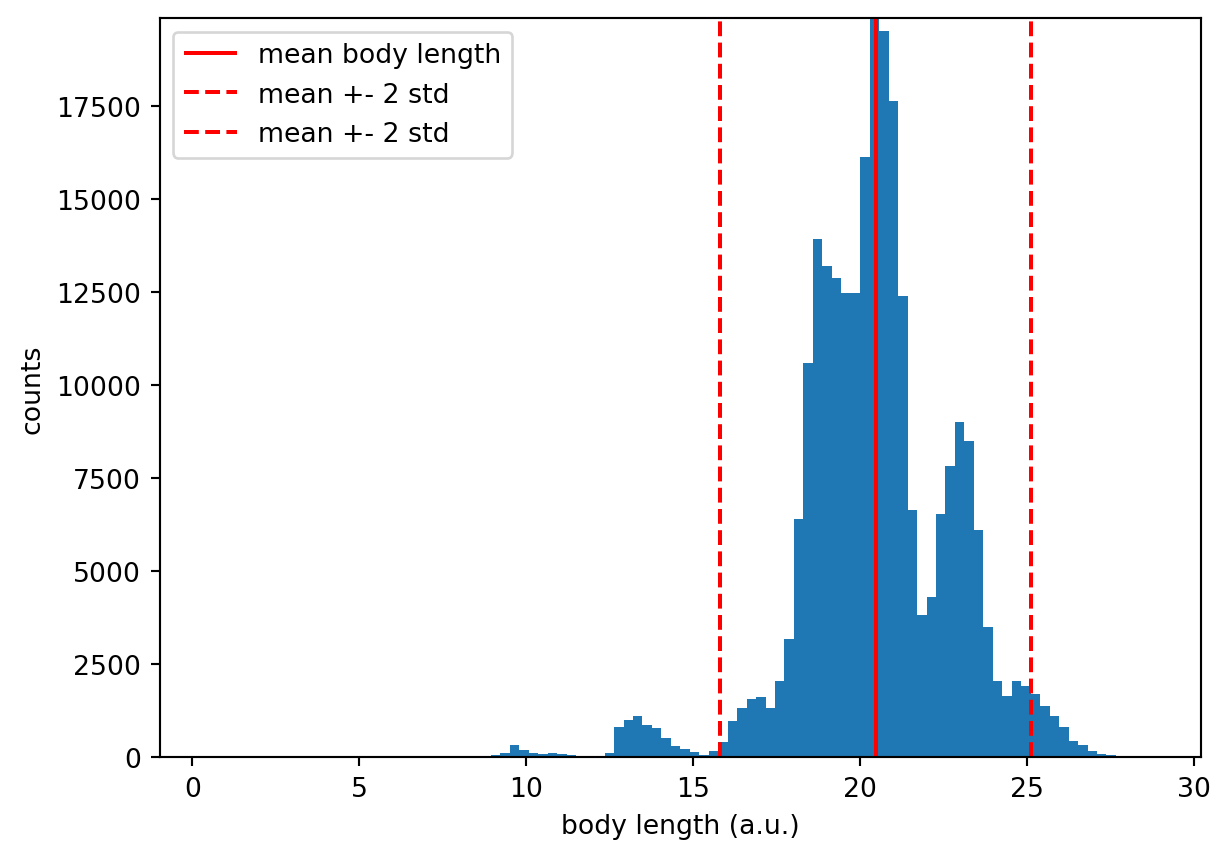
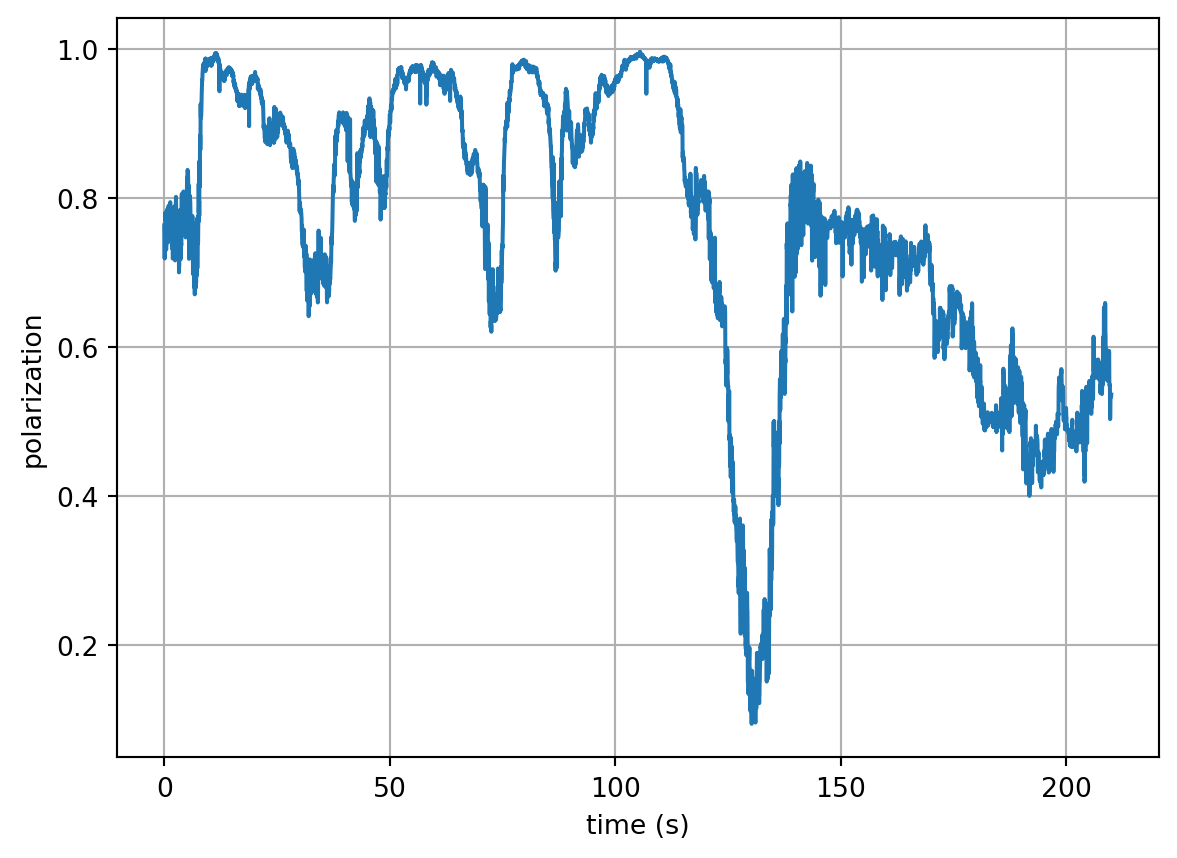
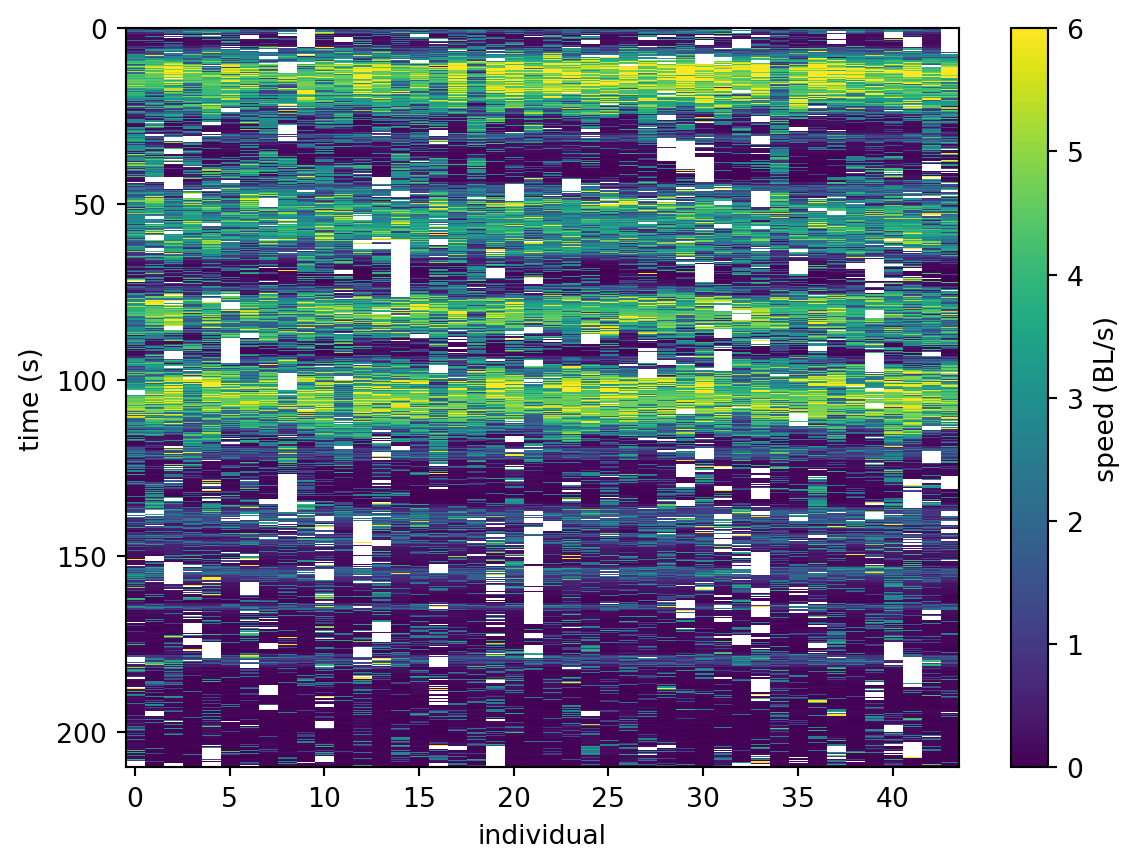
1.035e+03 1.043e+03 ... 680.6 743.0
array([[[[1035.0875 , 1042.7065 , 1305.2988 , ..., 1288.8965 ,
1075.2968 , nan],
[1055.7279 , 1056.1145 , 1287.899 , ..., 1272.5366 ,
1061.8053 , nan]],
[[1170.7526 , 1198.7987 , 1207.2275 , ..., 1196.4867 ,
1274.6531 , nan],
[1175.2821 , 1195.3256 , 1216.0687 , ..., 1210.1553 ,
1258.3647 , nan]]],
[[[1036.3914 , 1047.7836 , 1306.7142 , ..., 1290.4705 ,
1076.7327 , nan],
[1056.8123 , 1057.0919 , 1289.186 , ..., 1274.0607 ,
1062.8341 , nan]],
[[1170.8033 , 1197.9618 , 1207.1444 , ..., 1196.7869 ,
1274.542 , nan],
[1175.3003 , 1195.433 , 1216.0906 , ..., 1210.2795 ,
1258.3342 , nan]]],
...
5037.79 , 4924.202 ],
[ nan, 4916.7803 , nan, ..., 5216.492 ,
5016.4253 , 4944.2705 ]],
[[ nan, 731.70557, nan, ..., 994.94434,
689.243 , 738.3227 ],
[ nan, 731.16486, nan, ..., 975.09174,
680.5491 , 743.00867]]],
[[[ nan, 4897.603 , nan, ..., 5213.083 ,
5037.834 , 4924.2344 ],
[ nan, 4916.813 , nan, ..., 5216.505 ,
5016.4824 , 4944.314 ]],
[[ nan, 731.7324 , nan, ..., 994.9447 ,
689.2539 , 738.3456 ],
[ nan, 731.19904, nan, ..., 975.0833 ,
680.563 , 743.02875]]]],
shape=(6294, 2, 2, 44), dtype=float32)