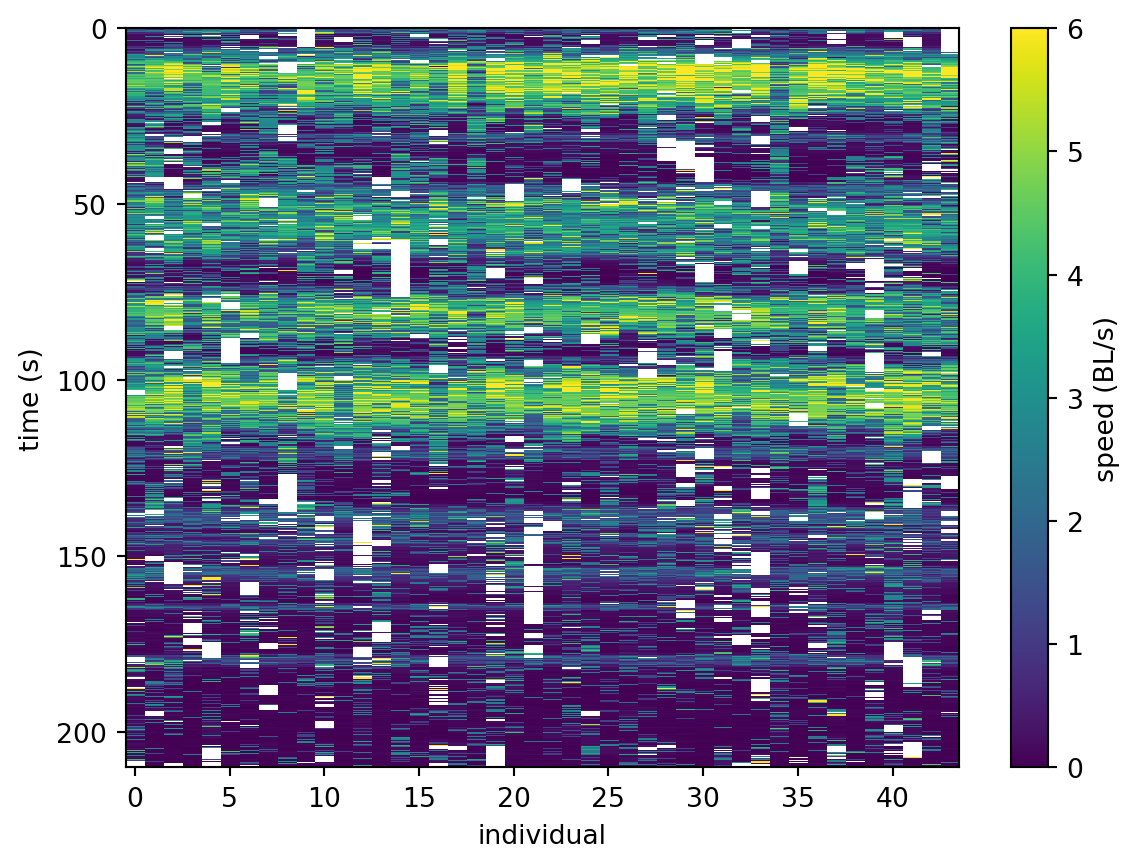
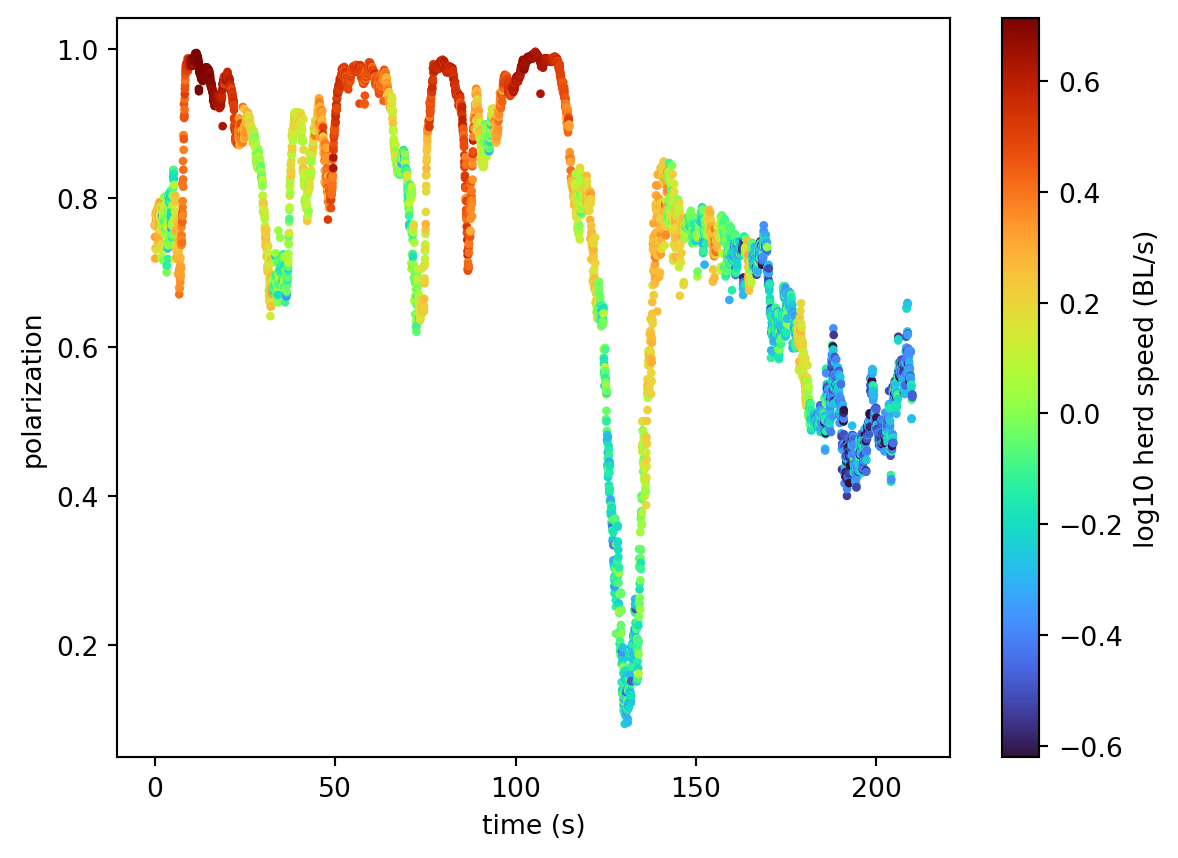
Downloading file 'poses/SLEAP_OSFM_zebras_drone.h5' from 'https://gin.g-node.org/neuroinformatics/movement-test-data/raw/master/poses/SLEAP_OSFM_zebras_drone.h5' to '/home/runner/.movement/data'.
0.00B [00:00, ?B/s]9.22kB [00:00, 62.5kB/s]49.2kB [00:00, 183kB/s] 89.1kB [00:00, 183kB/s]171kB [00:00, 310kB/s] 213kB [00:00, 300kB/s]245kB [00:00, 271kB/s]305kB [00:01, 309kB/s]359kB [00:01, 328kB/s]397kB [00:01, 304kB/s]439kB [00:01, 297kB/s]473kB [00:01, 275kB/s]515kB [00:01, 276kB/s]565kB [00:02, 293kB/s]608kB [00:02, 291kB/s]649kB [00:02, 285kB/s]695kB [00:02, 292kB/s]737kB [00:02, 288kB/s]782kB [00:02, 291kB/s]828kB [00:02, 295kB/s]858kB [00:03, 205kB/s]925kB [00:03, 264kB/s]955kB [00:03, 217kB/s]998kB [00:03, 234kB/s]1.03MB [00:03, 223kB/s]1.05MB [00:04, 207kB/s]1.08MB [00:04, 184kB/s]1.11MB [00:04, 181kB/s]1.13MB [00:04, 153kB/s]1.18MB [00:04, 179kB/s]1.21MB [00:05, 177kB/s]1.23MB [00:05, 173kB/s]1.26MB [00:05, 171kB/s]1.29MB [00:05, 170kB/s]1.31MB [00:05, 171kB/s]1.34MB [00:05, 169kB/s]1.37MB [00:05, 184kB/s]1.40MB [00:06, 185kB/s]1.43MB [00:06, 193kB/s]1.45MB [00:06, 184kB/s]1.49MB [00:06, 195kB/s]1.51MB [00:06, 188kB/s]1.55MB [00:06, 199kB/s]1.57MB [00:06, 189kB/s]1.60MB [00:07, 199kB/s]1.64MB [00:07, 212kB/s]1.67MB [00:07, 203kB/s]1.70MB [00:07, 207kB/s]1.73MB [00:07, 210kB/s]1.77MB [00:07, 224kB/s]1.80MB [00:08, 224kB/s]1.83MB [00:08, 222kB/s]1.87MB [00:08, 228kB/s]1.91MB [00:08, 239kB/s]1.95MB [00:08, 253kB/s]1.99MB [00:08, 248kB/s]2.03MB [00:08, 254kB/s]2.07MB [00:09, 266kB/s]2.11MB [00:09, 266kB/s]2.15MB [00:09, 272kB/s]2.20MB [00:09, 274kB/s]2.24MB [00:09, 288kB/s]2.29MB [00:09, 291kB/s]2.33MB [00:09, 288kB/s]2.38MB [00:10, 292kB/s]2.42MB [00:10, 300kB/s]2.48MB [00:10, 310kB/s]2.52MB [00:10, 301kB/s]2.57MB [00:10, 340kB/s]2.60MB [00:10, 337kB/s]2.64MB [00:10, 345kB/s]2.69MB [00:11, 344kB/s]2.75MB [00:11, 406kB/s]2.79MB [00:11, 399kB/s]2.83MB [00:11, 394kB/s]2.89MB [00:11, 415kB/s]2.96MB [00:11, 470kB/s]3.00MB [00:11, 461kB/s]3.07MB [00:11, 494kB/s]3.13MB [00:11, 511kB/s]3.22MB [00:12, 424kB/s]3.36MB [00:12, 607kB/s]3.43MB [00:12, 520kB/s]3.48MB [00:12, 523kB/s]3.54MB [00:12, 412kB/s]3.59MB [00:12, 387kB/s]3.63MB [00:13, 357kB/s]3.67MB [00:13, 290kB/s]3.70MB [00:13, 271kB/s]3.73MB [00:13, 250kB/s]3.76MB [00:13, 230kB/s]3.79MB [00:13, 211kB/s]3.82MB [00:14, 209kB/s]3.85MB [00:14, 212kB/s]3.88MB [00:14, 201kB/s]3.91MB [00:14, 208kB/s]3.94MB [00:14, 207kB/s]3.98MB [00:14, 220kB/s]4.01MB [00:15, 215kB/s]4.04MB [00:15, 210kB/s]4.07MB [00:15, 214kB/s]4.09MB [00:15, 166kB/s]4.13MB [00:15, 196kB/s]4.15MB [00:15, 198kB/s]4.17MB [00:15, 171kB/s]4.20MB [00:16, 187kB/s]4.23MB [00:16, 180kB/s]4.26MB [00:16, 195kB/s]4.29MB [00:16, 191kB/s]4.33MB [00:16, 211kB/s]4.36MB [00:16, 217kB/s]4.39MB [00:16, 213kB/s]4.43MB [00:17, 215kB/s]4.47MB [00:17, 218kB/s]4.50MB [00:17, 222kB/s]4.53MB [00:17, 221kB/s]4.56MB [00:17, 232kB/s]4.60MB [00:17, 228kB/s]4.64MB [00:18, 242kB/s]4.67MB [00:18, 234kB/s]4.70MB [00:18, 229kB/s]4.74MB [00:18, 210kB/s]4.77MB [00:18, 228kB/s]4.81MB [00:18, 223kB/s]4.84MB [00:18, 240kB/s]4.88MB [00:19, 231kB/s]4.91MB [00:19, 196kB/s]4.96MB [00:19, 250kB/s]4.98MB [00:19, 231kB/s]5.01MB [00:19, 221kB/s]5.05MB [00:19, 220kB/s]5.08MB [00:20, 219kB/s]0.00B [00:00, ?B/s] 0.00B [00:00, ?B/s]